Explore our protein model - can you guess the protein?
Bringing AI protein design to students
DLAI is a student association at EPFL dedicated to designing proteins with AI and computational tools, then testing them in the lab to see if the engineering really works.
We create new binders, metal‑binding proteins and immune‑targeted designs in collaboration with research labs across campus, turning cutting‑edge methods in protein engineering into hands‑on projects for students.
Our mission
We aim to bridge academia and student experience by connecting undergraduates and graduate students with EPFL labs that support our projects, offer guidance and propose real design challenges.
Through workshops, design sprints and MAKE projects, we explore how AI‑based models and physical experiments can work together to build useful, creative and responsible new proteins.
The people behind DLAI
We are engineering, life‑science and computer‑science students with a shared interest in protein design, synthetic biology and AI.
Cris Darbellay
Cris Darbellay
Leads the association, coordinates partners and keeps DLAI aligned with the MAKE and EPFL communities.
Rémy Mühlethaler
Rémy Mühlethaler
Organises the team, represents the association in official events, and manages internal communications.
Alexia Océane Möller
Alexia Océane Möller
Oversees project pipelines from AI design to wet‑lab validation, and interfaces with partner labs.
Emma S. Castelli
Emma S. Castelli
Organises workshops, events and the Protein Design Week, and manages web development for DLAI.
Nicolas Pafumi
Nicolas Pafumi
Manages the association's finances and ensures proper budget allocation for projects and events.
Elsa Sànchez Fernàndez
Elsa Sànchez Fernàndez
Manages communications and social media for DLAI, ensuring consistent messaging and engagement with the community.
Tolga Semiz
Tolga Semiz
Team lead of the Cultivated Meat project, coordinating design, experiments and collaboration with partner labs.
Designing, building and testing proteins
Our projects span AI‑driven sequence design, structure prediction, and experimental validation in collaboration with EPFL labs. Here you can keep track of ongoing work and also calls for open projects.
Ongoing Projects
Phosphosite engineering
This project combines molecular biology and artificial intelligence to investigate conformational changes in phosphosites upon phosphorylation, analyzing over 20,000 phosphosites using custom Python scripts and refined selections with Alphafold predictions. The next phase involves validating about 100 selected phosphosites using NMR spectroscopy to confirm AI model predictions, potentially advancing our understanding of protein functionality and leading to groundbreaking applications in pharmaceuticals and biomolecular studies.
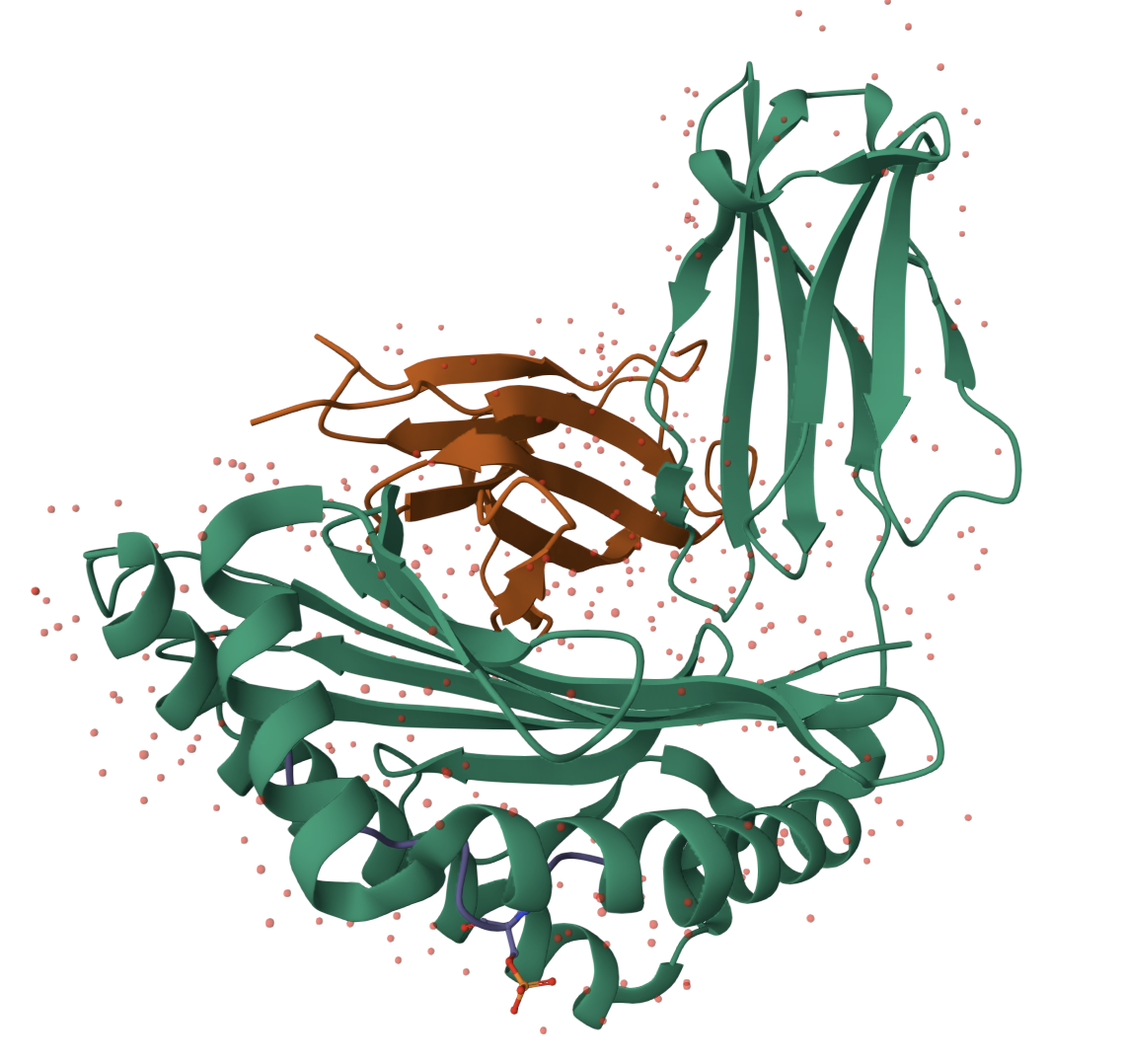
Light Activated Kinases
We are trying to modify protein kinases by removing some of their domains but such that they keep their usual functionality in cells. Once we have these we’d like to fuse them to other proteins/ protein domains to be able to better regulate the protein’s activity (eg. addition of a light activating domain so that light would act as an on/off switch).
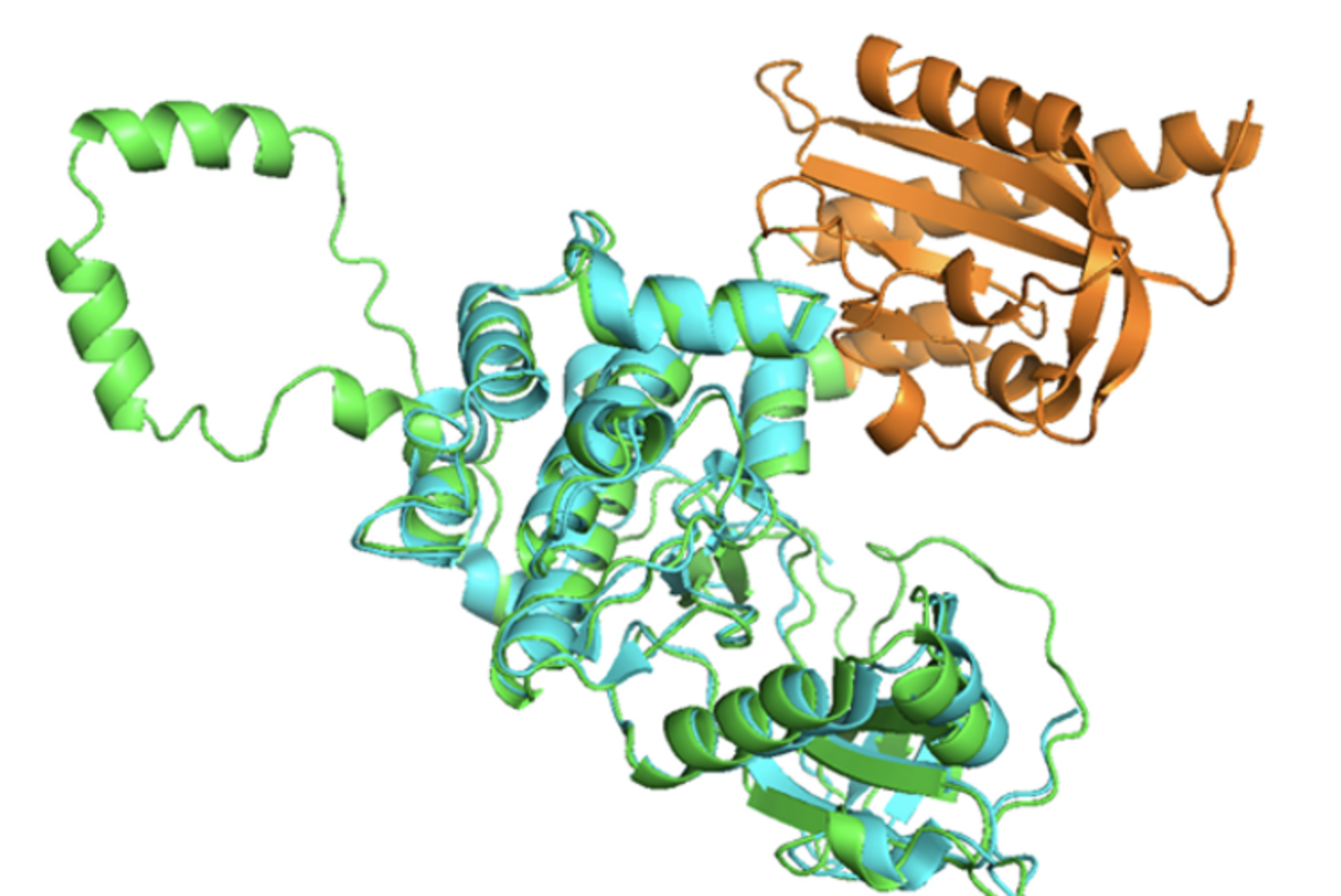
Hydrophobic mutants of proteins
The goal was to to find hydrophobic mutants of proteins while keeping the shape by randomized mutations. The hydrophilic amino acids on the surface have been replaced by hydrophobic ones.
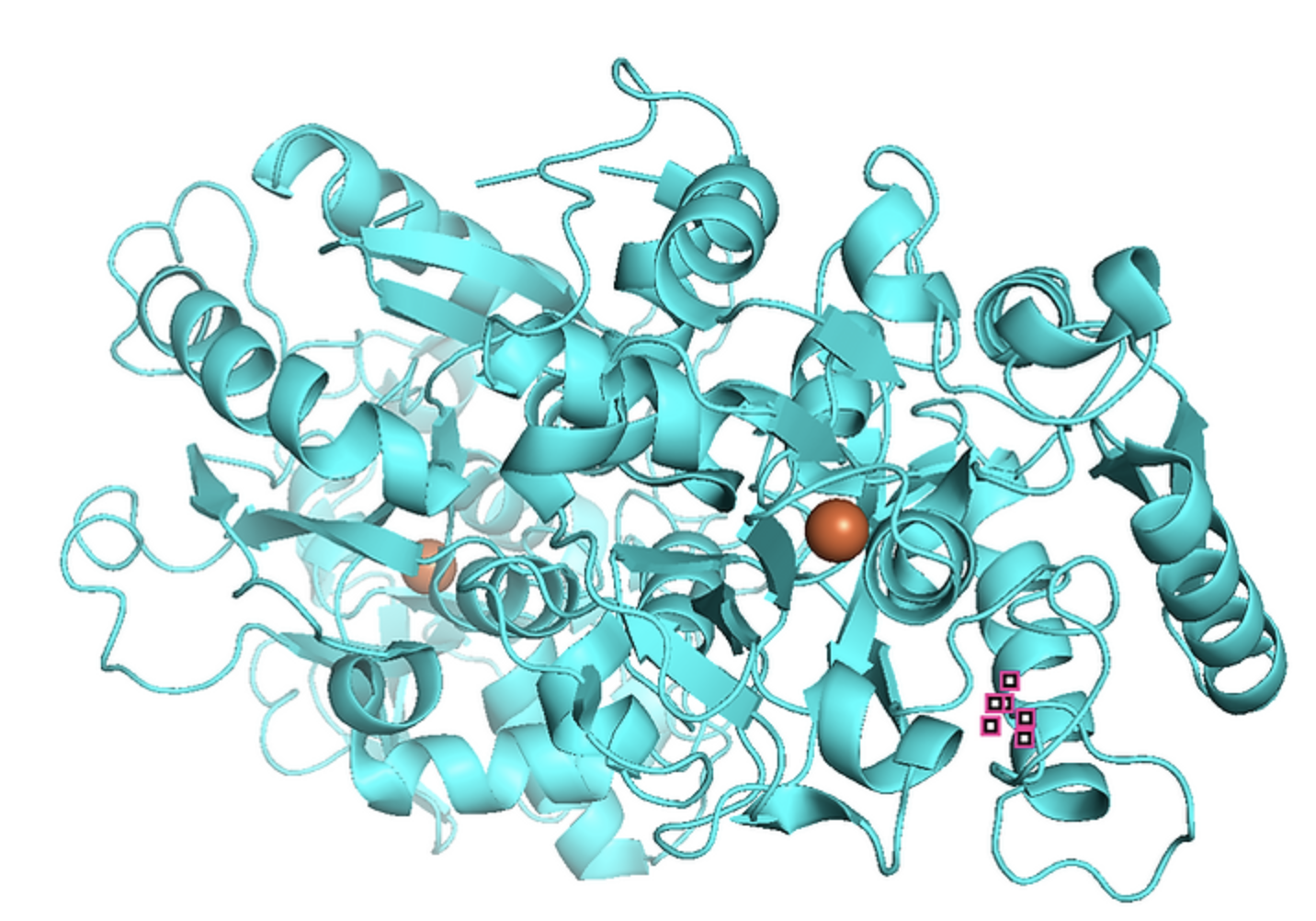
Designing and testing EphA2 receptor binders
The goal is to design and test binders for EphA2 receptors, which are receptors overexpressed in tumoral cells.
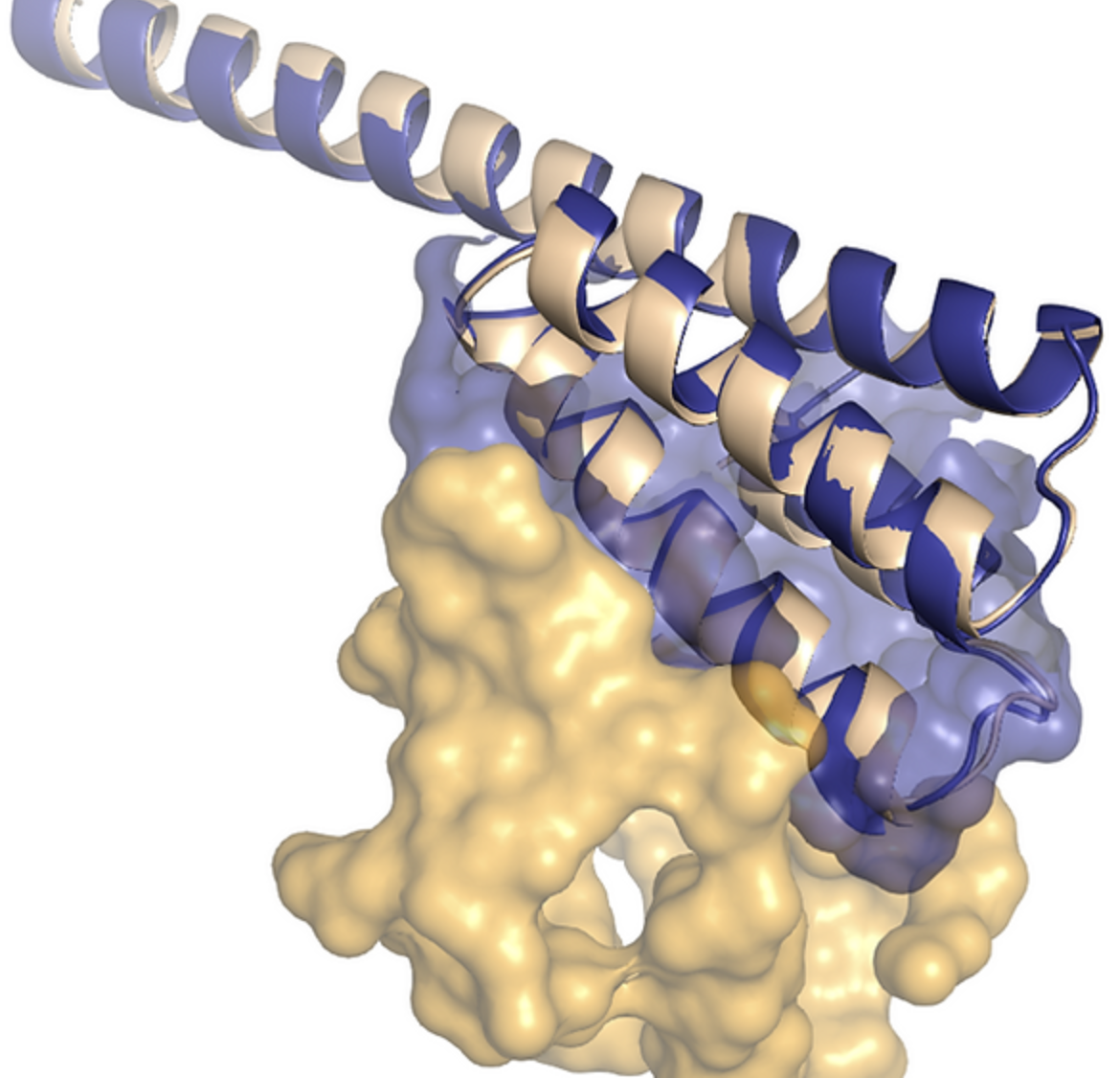
Introducing an alternative alcoholic fermentation pathway in yeast
We are four life sciences engineering students working on optimizing ethanol production through yeast anaerobic fermentation. Our goal is to establish an alternative fermentation pathway in yeasts by studying the integration of a bacterial fermentation pathway presenting advantages such as alleviating anaerobic redox stress or bypassing toxic intermediates. The efficiency of the enzymes involved will then be optimised using novel AI tools such as RFdiffusion.
[URE3] prion binder
We want to create a protein binder that binds to the [URE3] prion in S. cerevisiae, with the future goal of being able to design a binder for prions causing diseases in humans.
Development of a Novel Probe-Free Non-Invasive Imaging Method for Intracellular Antigens
This project aims at developing a novel non-invasive, probe-free method for imaging intracellular antigens.
GPCR Biosensor Development: Advancing Molecular Detection via BRET and Synthetic Biology
The GPCR Biosensor project focuses on detecting the activation of Gαi/t-subunits in yeast through GPCRs using a BRET (Bioluminescence Resonance Energy Transfer) system. Our current method involves the KB1753 peptide, acting as a proof of concept in laboratory conditions. The aim is to develop this system to recognize a range of molecules, like histamine, by modifying the GPCR binding site. This could lead to significant advancements in understanding signal transduction and drug interactions.The team's efforts are grounded in seminal research, with foundational work by C. Johnston et al. in Biochemistry, 2006, and R. Jefferson et al. in Nature, 2023, focusing on the minimal determinants for Gαi/t binding and the dynamic receptor-peptide signaling complexes, respectively. This project represents a convergence of molecular biology, biochemistry, and synthetic biology, aiming to create a versatile tool for molecular detection and study.
Partner labs & supporters
DLAI works closely with EPFL research groups and external partners who provide scientific guidance, lab space and design challenges. Here is an overview of our hosting laboratories.
Our projects are developed in collaboration with EPFL research groups who provide scientific guidance, lab space and design challenges. Here is an overview of our hosting laboratories.
To see all open projects click HERE
Sahand Rahi lab
Prof Rahi's lab focuses on Timing of Cellular Decisions, Directed Evolution, AI- and Physics-based Design of Computational Protein Functionalities, Neural Computation Across Timescales, and Neural Computation Across Timescales.

Dal Peraro - Molecular Biomodelling Lab
The main goal of Dal Peraro's laboratory is to understand the physical and chemical properties of complex biological systems, in particular their function emerging from structure. To address these questions, we use and develop a broad spectrum of computational tools fully integrated with experimental data. Multiscale models and dynamic integrative modeling are used to investigate the assembly and function of molecular assemblies mimicking conditions of the cellular environment.

Correia - Protein Design and Immunoengineering lab
Correia's group is driven by the passion of expanding nature’s repertoire by computationally designing novel functional proteins to be used for practical purposes such as therapeutics, vaccines, biosensors and others.

Barth - Laboratory of Protein and Cell Engineering
The LPCE works at the interface of biophysics, chemical, structural, computational and cell biology to uncover the molecular principles that regulate protein and cellular signaling. We use this understanding to (1) design protein systems with novel biosensing and signaling functions for synthetic biology and engineered cell therapeutic applications; (2) predict the effects of genetic variations on protein structure/function for personalized cancer medicine applications.

De Los Rios - Laboratory of Statistical biophysics
The Laboratory of Statistical Biophysics (or Laboratoire de Biophysique Statistique, if you like the sound of French), has been established in October 2003. Our offices are located in the stimulating scientific environment of the EPFL/UNIL campus, and with a stunning view on the lake. Our goal is to use tools mainly from statistical physics to contribute to the understanding of the properties of living matter. In general, we are interested in statistical physics, biophysics and computational physics.

Become a sponsor
We are still looking for partners and sponsors who can support us.

Stay connected
Before joining any projects, please carefully read the expectations .
For any supplementary information on this MAKE project, please contact one of the following: dlai@epfl.ch, cris.darbellay@epfl.ch, remy.mulhethaler@epfl.ch, alexia.moller@epfl.ch, emma.castelli@epfl.ch
For any information about a specific project, please write an email to dlai@epfl.ch, and cc one person from the above addresses. You can find project names, labs, and info on open projects.
If you are interested in our project, please join our whatsapp channel for updates.
Follow our activities, events and project updates on social media and EPFL channels.

